smriprep.workflows.fit.registration module
Spatial normalization workflows.
- smriprep.workflows.fit.registration.init_register_template_wf(*, sloppy: bool, omp_nthreads: int, templates: list[str], image_type: str = 'T1w', name='register_template_wf')[source]
Build an individual spatial normalization workflow using
antsRegistration.- Workflow Graph
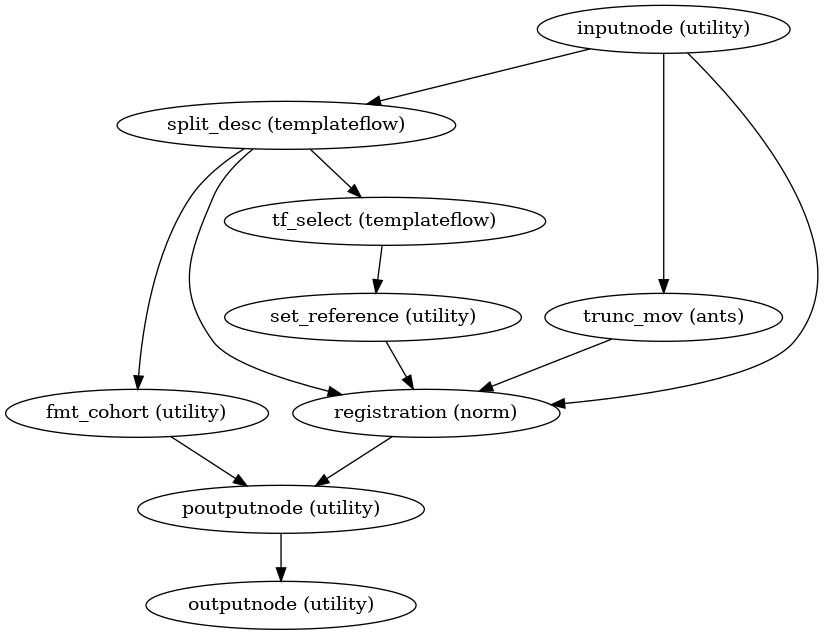
(Source code, png, svg, pdf)
Important
This workflow defines an iterable input over the input parameter
templates, so Nipype will produce one copy of the downstream workflows which connectpoutputnode.templateorpoutputnode.template_specto their inputs (poutputnodestands for parametric output node). Nipype refers to this expansion of the graph as parameterized execution. If a joint list of values is required (and thus cutting off parameterization), please use the equivalent outputs ofoutputnode(which joins all the parameterized execution paths).- Parameters:
sloppy (
bool) – Apply sloppy arguments to speed up processing. Use with caution, registration processes will be very inaccurate.omp_nthreads (
int) – Maximum number of threads an individual process may use.templates (
listofstr) – List of standard space fullnames (e.g.,MNI152NLin6AsymorMNIPediatricAsym:cohort-4) which are targets for spatial normalization.
- Inputs:
moving_image – The input image that will be normalized to standard space.
lesion_mask – (optional) A mask to exclude regions from the cost-function input domain to enable standardization of lesioned brains.
template – Template name and specification
image_type – Moving image modality
- Outputs:
anat2std_xfm – The T1w-to-template transform.
std2anat_xfm – The template-to-T1w transform.
template – Template name extracted from the input parameter
template, for further use in downstream nodes.template_spec – Template specifications extracted from the input parameter
template, for further use in downstream nodes.