niworkflows.anat.ants module¶
Nipype translation of ANTs’ workflows.
- niworkflows.anat.ants.init_atropos_wf(name='atropos_wf', use_random_seed=True, omp_nthreads=None, mem_gb=3.0, padding=10, in_segmentation_model=(3, 1, 2, 3), bspline_fitting_distance=200, wm_prior=False)[source]¶
Create an ANTs’ ATROPOS workflow for brain tissue segmentation.
Re-interprets supersteps 6 and 7 of
antsBrainExtraction.sh, which refine the mask previously computed with the spatial normalization to the template. The workflow also executes steps 8 and 9 of the brain extraction workflow.- Workflow Graph
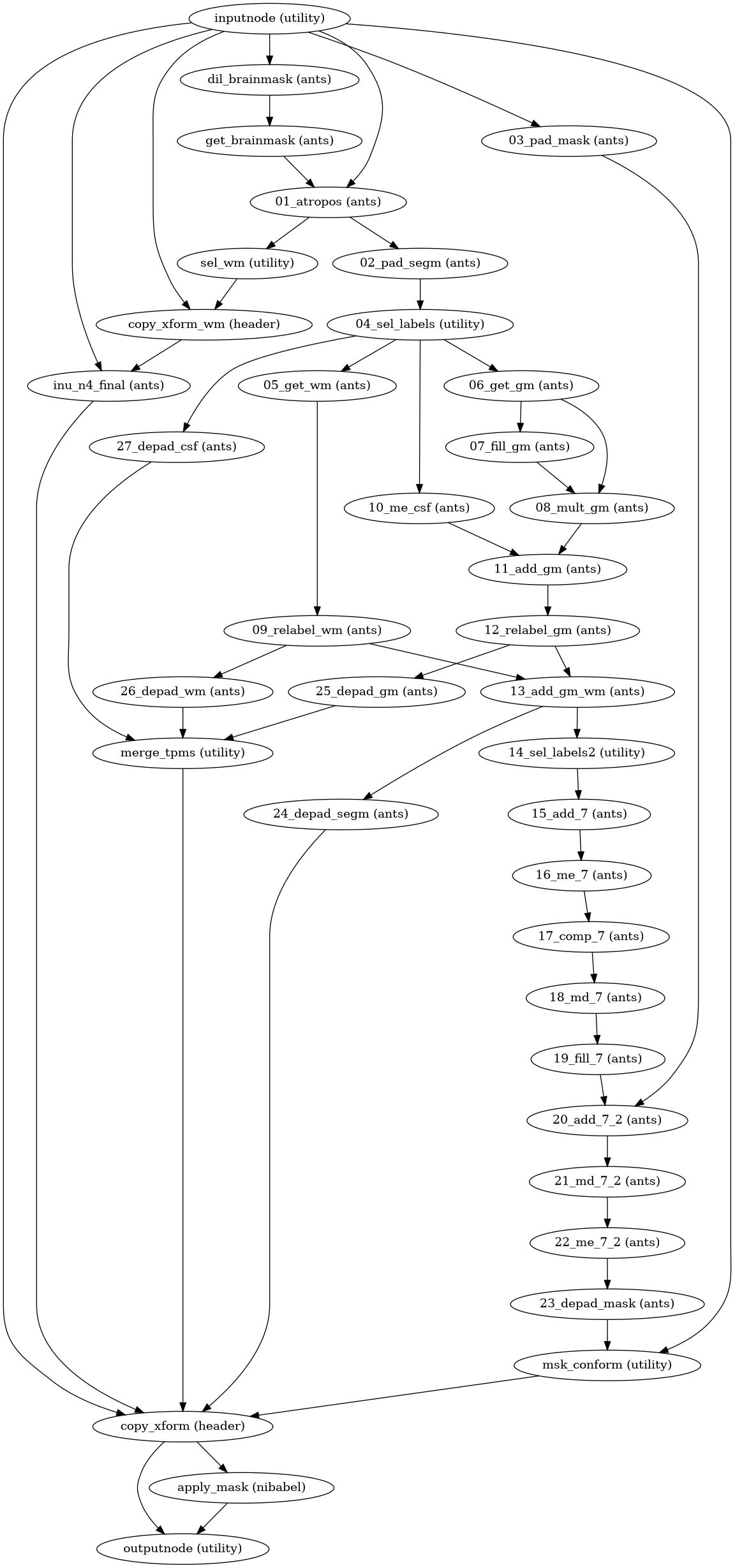
(Source code, png, svg, pdf)
- Parameters:
name (str, optional) – Workflow name (default: “atropos_wf”).
use_random_seed (bool) – Whether ATROPOS should generate a random seed based on the system’s clock
omp_nthreads (int) – Maximum number of threads an individual process may use
mem_gb (float) – Estimated peak memory consumption of the most hungry nodes in the workflow
padding (int) – Pad images with zeros before processing
in_segmentation_model (tuple) – A k-means segmentation is run to find gray or white matter around the edge of the initial brain mask warped from the template. This produces a segmentation image with \($K$\) classes, ordered by mean intensity in increasing order. With this option, you can control \($K$\) and tell the script which classes represent CSF, gray and white matter. Format (K, csfLabel, gmLabel, wmLabel). Examples:
(3,1,2,3)for T1 with K=3, CSF=1, GM=2, WM=3 (default),(3,3,2,1)for T2 with K=3, CSF=3, GM=2, WM=1,(3,1,3,2)for FLAIR with K=3, CSF=1 GM=3, WM=2,(4,4,2,3)uses K=4, CSF=4, GM=2, WM=3.bspline_fitting_distance (float) – The size of the b-spline mesh grid elements, in mm (default: 200)
wm_prior (
bool) – Whether the WM posterior obtained with ATROPOS should be regularized with a prior map (typically, mapped from the template). Whenwm_priorisTruethe input fieldwm_priorof the input node must be connected.
- Inputs:
in_files (list) – The original anatomical images passed in to the brain-extraction workflow.
in_corrected (list) – INU-corrected files.
in_mask (str) – Brain mask calculated previously.
wm_prior (
str) – Path to the WM prior probability map, aligned with the individual data.
- Outputs:
out_file (
str) – Path of the corrected and brain-extracted result, using the ATROPOS refinement.bias_corrected (
str) – Path of the corrected and result, using the ATROPOS refinement.bias_image (
str) – Path of the estimated INU bias field, using the ATROPOS refinement.out_mask (str) – Refined brain mask
out_segm (str) – Output segmentation
out_tpms (str) – Output TPMs
- niworkflows.anat.ants.init_brain_extraction_wf(name='brain_extraction_wf', in_template='OASIS30ANTs', template_spec=None, use_float=True, normalization_quality='precise', omp_nthreads=None, mem_gb=3.0, bids_suffix='T1w', atropos_refine=True, atropos_use_random_seed=True, atropos_model=None, use_laplacian=True, bspline_fitting_distance=200)[source]¶
Build a workflow for atlas-based brain extraction on anatomical MRI data.
This is a Nipype implementation of atlas-based brain extraction inspired by the official ANTs’
antsBrainExtraction.shworkflow (only for 3D images).The workflow follows the following structure:
Step 1 performs several clerical tasks (preliminary INU correction, calculating the Laplacian of inputs, affine initialization) and the core spatial normalization.
Maps the brain mask into target space using the normalization calculated in 1.
Superstep 1b: binarization of the brain mask
Maps the WM (white matter) probability map from the template, if such prior exists. Combines the BS (brainstem) probability map before mapping if the WM and BS are given separately (as it is the case for
OASIS30ANTs.)Run a second N4 INU correction round, using the prior mapped into individual step in step 4 if available.
Superstep 6: apply ATROPOS on the INU-corrected result of step 5, and massage its outputs
Superstep 7: use results from 4 to refine the brain mask
If exist, use priors from step 4, calculate the overlap of the posteriors estimated in step 4 to select that overlapping the most with the WM+BS prior from the template. Combine that posterior with the refined brain mask and pass it on to the next step.
Apply a final N4 using the refined brain mask (or the map calculated in step 8 if priors were found) as weights map for the algorithm.
- Workflow Graph
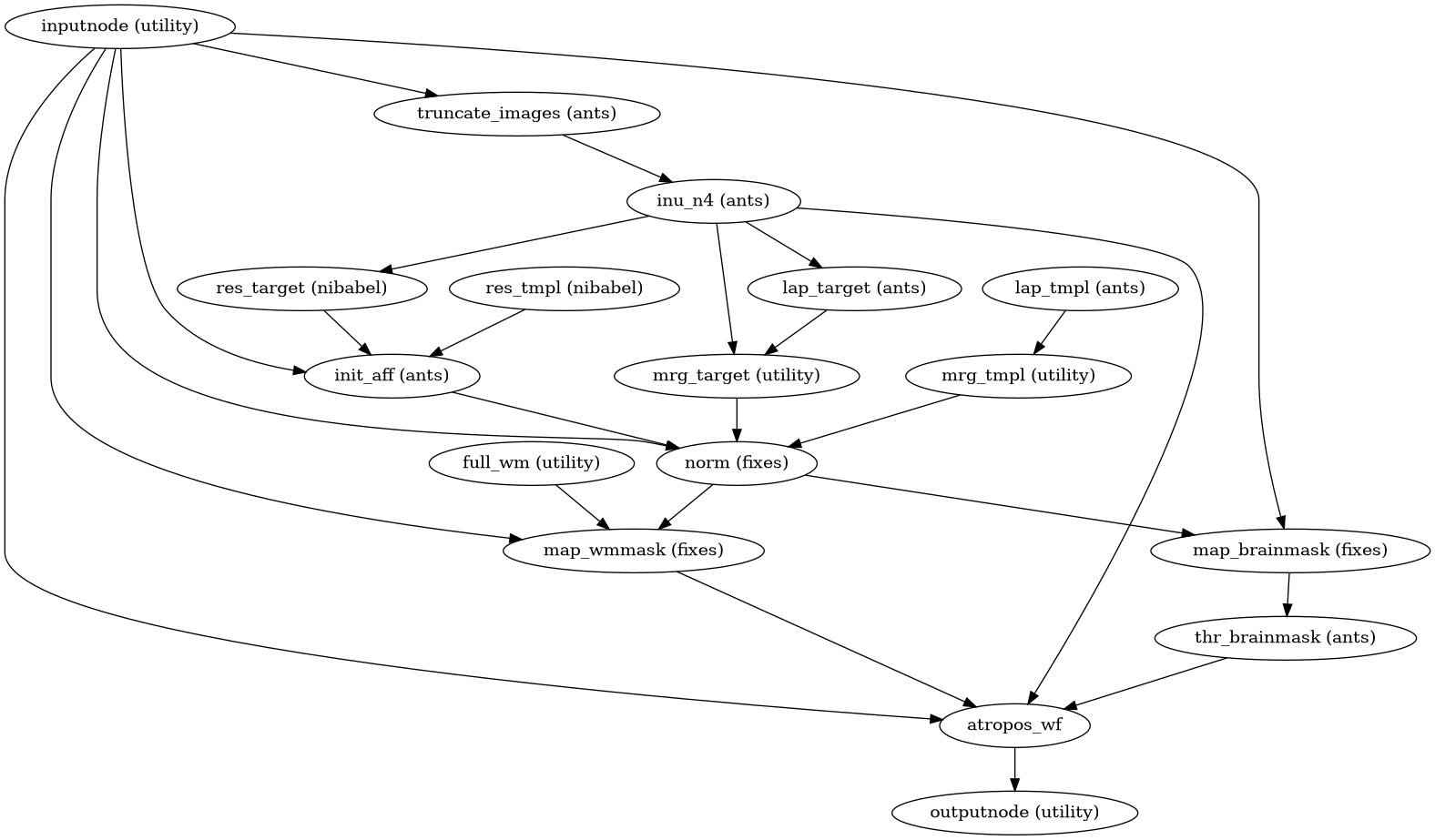
(Source code, png, svg, pdf)
- Parameters:
in_template (str) – Name of the skull-stripping template (‘OASIS30ANTs’, ‘NKI’, or path). The brain template from which regions will be projected Anatomical template created using e.g. LPBA40 data set with
buildtemplateparallel.shin ANTs. The workflow will automatically search for a brain probability mask created using e.g. LPBA40 data set which have brain masks defined, and warped to anatomical template and averaged resulting in a probability image.use_float (bool) – Whether single precision should be used
normalization_quality (str) – Use more precise or faster registration parameters (default:
precise, other possible values:testing)omp_nthreads (int) – Maximum number of threads an individual process may use
mem_gb (float) – Estimated peak memory consumption of the most hungry nodes in the workflow
bids_suffix (str) – Sequence type of the first input image. For a list of acceptable values see https://bids-specification.readthedocs.io/en/latest/04-modality-specific-files/01-magnetic-resonance-imaging-data.html#anatomy-imaging-data
atropos_refine (bool) – Enables or disables the whole ATROPOS sub-workflow
atropos_use_random_seed (bool) – Whether ATROPOS should generate a random seed based on the system’s clock
atropos_model (tuple or None) – Allows to specify a particular segmentation model, overwriting the defaults based on
bids_suffixuse_laplacian (bool) – Enables or disables alignment of the Laplacian as an additional criterion for image registration quality (default: True)
bspline_fitting_distance (float) – The size of the b-spline mesh grid elements, in mm (default: 200)
name (str, optional) – Workflow name (default: antsBrainExtraction)
- Inputs:
in_files (list) – List of input anatomical images to be brain-extracted, typically T1-weighted. If a list of anatomical images is provided, subsequently specified images are used during the segmentation process. However, only the first image is used in the registration of priors. Our suggestion would be to specify the T1w as the first image.
in_mask (list, optional) – Mask used for registration to limit the metric computation to a specific region.
- Outputs:
out_file (str) – Skull-stripped and INU-corrected
in_filesout_mask (str) – Calculated brain mask
bias_corrected (str) – The
in_filesinput images, after INU correction, before skull-stripping.bias_image (str) – The INU field estimated for each input in
in_filesout_segm (str) – Output segmentation by ATROPOS
out_tpms (str) – Output TPMs by ATROPOS
- niworkflows.anat.ants.init_n4_only_wf(atropos_model=None, atropos_refine=True, atropos_use_random_seed=True, bids_suffix='T1w', mem_gb=3.0, name='n4_only_wf', omp_nthreads=None)[source]¶
Build a workflow to sidetrack brain extraction on skull-stripped datasets.
An alternative workflow to “init_brain_extraction_wf”, for anatomical images which have already been brain extracted.
Creates brain mask assuming all zero voxels are outside the brain
Applies N4 bias field correction
(Optional) apply ATROPOS and massage its outputs
Use results from 3 to refine N4 bias field correction
- Workflow Graph
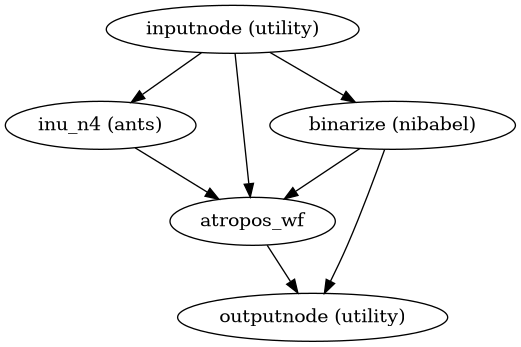
(Source code, png, svg, pdf)
- Parameters:
omp_nthreads (int) – Maximum number of threads an individual process may use
mem_gb (float) – Estimated peak memory consumption of the most hungry nodes
bids_suffix (str) – Sequence type of the first input image. For a list of acceptable values see https://bids-specification.readthedocs.io/en/latest/04-modality-specific-files/01-magnetic-resonance-imaging-data.html#anatomy-imaging-data
atropos_refine (bool) – Enables or disables the whole ATROPOS sub-workflow
atropos_use_random_seed (bool) – Whether ATROPOS should generate a random seed based on the system’s clock
atropos_model (tuple or None) – Allows to specify a particular segmentation model, overwriting the defaults based on
bids_suffixname (str, optional) – Workflow name (default:
'n4_only_wf').
- Inputs:
in_files – List of input anatomical images to be bias corrected, typically T1-weighted. If a list of anatomical images is provided, subsequently specified images are used during the segmentation process. However, only the first image is used in the registration of priors. Our suggestion would be to specify the T1w as the first image.
- Outputs:
out_file – INU-corrected
in_filesout_mask – Calculated brain mask
bias_corrected – Same as “out_file”, provided for consistency with brain extraction
bias_image – The INU field estimated for each input in
in_filesout_segm – Output segmentation by ATROPOS
out_tpms – Output TPMs by ATROPOS