sdcflows.workflows.fit.syn module¶
Estimating the susceptibility distortions without fieldmaps.
- sdcflows.workflows.fit.syn.init_syn_preprocessing_wf(*, atlas_threshold=3, debug=False, name='syn_preprocessing_wf', omp_nthreads=1, auto_bold_nss=False, t1w_inversion=False, sd_prior=True)[source]¶
Prepare EPI references and co-registration to anatomical for SyN.
- Workflow Graph
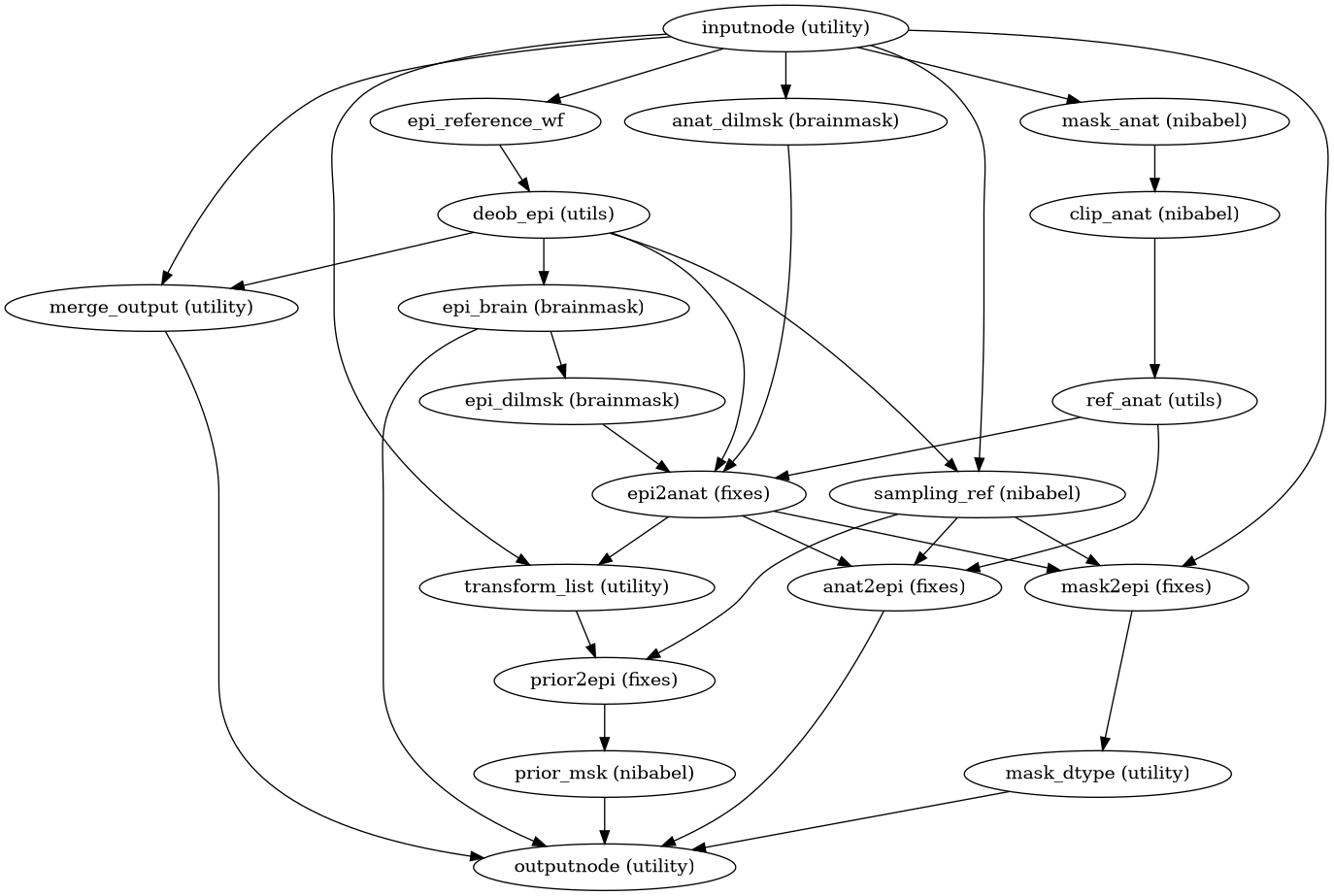
(Source code, png, svg, pdf)
- Parameters:
atlas_threshold (
float) – Mask excluding areas with average distortions below this threshold (in mm) on the prior.debug (
bool) – Whether a fast (less accurate) configuration of the workflow should be applied.name (
str) – Name for this workflowomp_nthreads (
int) – Parallelize internal tasks across the number of CPUs given by this option.auto_bold_nss (
bool) – Set up the reference workflow to automatically execute nonsteady states detection of BOLD images.t1w_inversion (
bool) – Run T1w intensity inversion so that it looks more like a T2 contrast.sd_prior (
bool) – Enable using a prior map to regularize the SyN cost function.
- Inputs:
in_epis (
listofstr) – Distorted EPI images that will be merged together to create the EPI reference file.t_masks (
listofbool) – (optional) mask of timepoints for calculating an EPI reference. Not used ifauto_bold_nss=True.in_meta (
listofdict) – Metadata dictionaries corresponding to thein_episinput.in_anat (
str) – A preprocessed anatomical (T1w or T2w) image.mask_anat (
str) – A brainmask corresponding to the anatomical (T1w or T2w) image.std2anat_xfm (
str) – inverse registration transform of T1w image to MNI template.
- Outputs:
epi_ref (
tuple(str,dict)) – A tuple, where the first element is the path of the distorted EPI reference map (e.g., an average of b=0 volumes), and the second element is a dictionary of associated metadata.anat_ref (
str) – Path to the anatomical, skull-stripped reference in EPI space.anat_mask (
str) – Path to the brain mask corresponding toanat_refin EPI space.sd_prior (
str) – A template map of areas with strong susceptibility distortions (SD) to regularize the cost function of SyN.
- sdcflows.workflows.fit.syn.init_syn_sdc_wf(*, use_metadata_estimates=False, fallback_total_readout_time=None, sloppy=False, debug=False, name='syn_sdc_wf', omp_nthreads=1, laplacian_weight=None, **kwargs)[source]¶
Build the fieldmap-less susceptibility-distortion estimation workflow.
SyN deformation is restricted to the phase-encoding (PE) direction. If no PE direction is specified, anterior-posterior PE is assumed.
- Workflow Graph
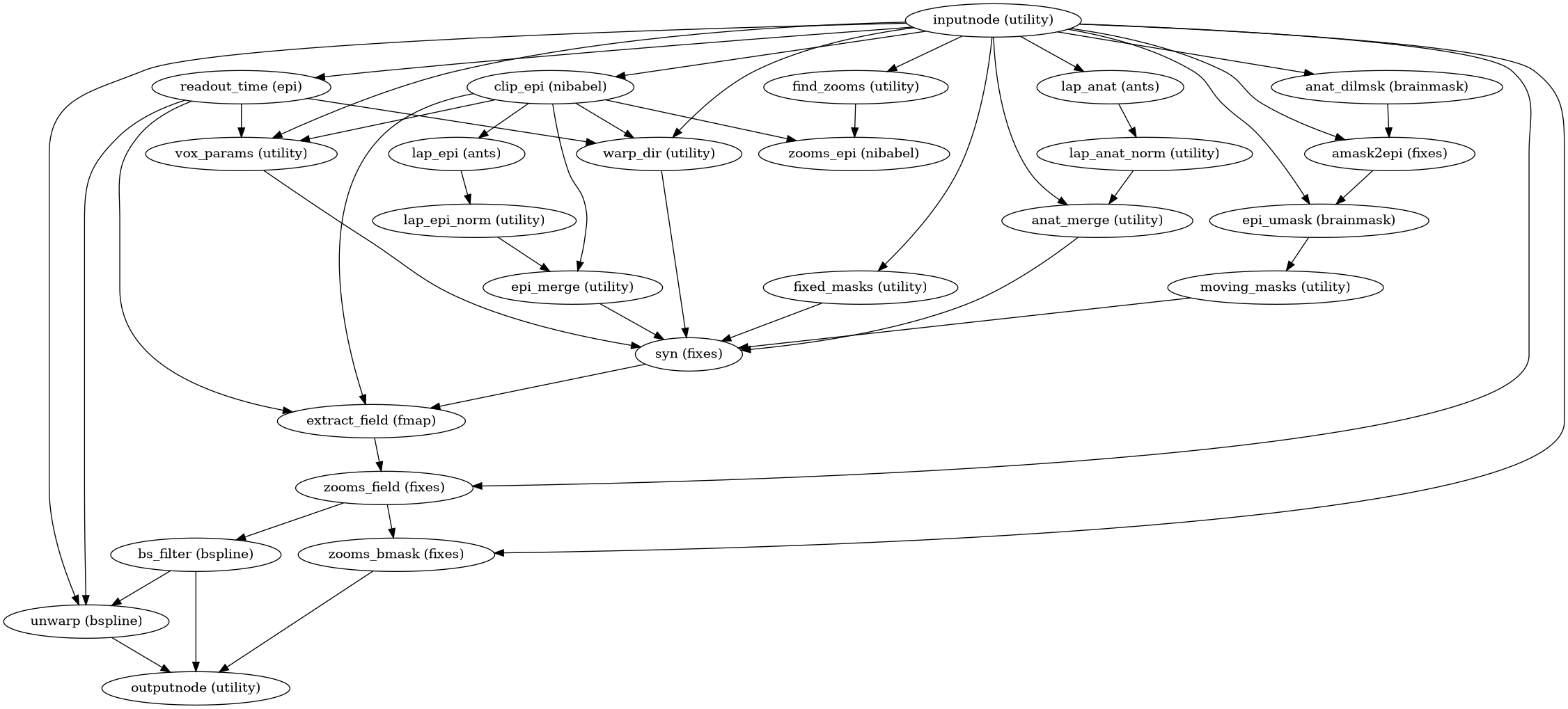
(Source code, png, svg, pdf)
- Parameters:
sloppy (
bool) – Whether a fast (less accurate) configuration of the workflow should be applied.debug (
bool) – Run in debug modename (
str) – Name for this workflowomp_nthreads (
int) – Parallelize internal tasks across the number of CPUs given by this option.laplacian_weight (
tuple(optional)) – Tuple of two weights of the Laplacian term in the SyN cost function (one weight per registration level).sd_prior (
bool) – Enable using a prior map to regularize the SyN cost function.
- Inputs:
epi_ref (
tuple(str,dict)) – A tuple, where the first element is the path of the distorted EPI reference map (e.g., an average of b=0 volumes), and the second element is a dictionary of associated metadata.epi_mask (
str) – A path to a brain mask corresponding toepi_ref.anat_ref (
str) – A preprocessed, skull-stripped anatomical (T1w or T2w) image resampled in EPI space.anat_mask (
str) – Path to the brain mask corresponding toanat_refin EPI space.
- Outputs:
fmap (
str) – The path of the estimated fieldmap.fmap_ref (
str) – The path of an unwarped conversion of files inepi_ref.fmap_coeff (
strorlistofstr) – The path(s) of the B-Spline coefficients supporting the fieldmap.out_warp (
str) – The path of the corresponding displacements field transform to unwarp susceptibility distortions.method (
str) – Short description of the estimation method that was run.